observation_id IgG_concentration age gender slum
1 5772 0.3176895 2 Female Non slum
2 8095 3.4368231 4 Female Non slum
3 9784 0.3000000 4 Male Non slumModule 7: Variable Creation, Classes, and Summaries
Learning Objectives
After module 7, you should be able to…
- Create new variables
- Characterize variable classes
- Manipulate the classes of variables
- Conduct 1 variable data summaries
Import data for this module
Let’s first read in the data from the previous module and look at it briefly with a new function head(). head() allows us to look at the first n observations.
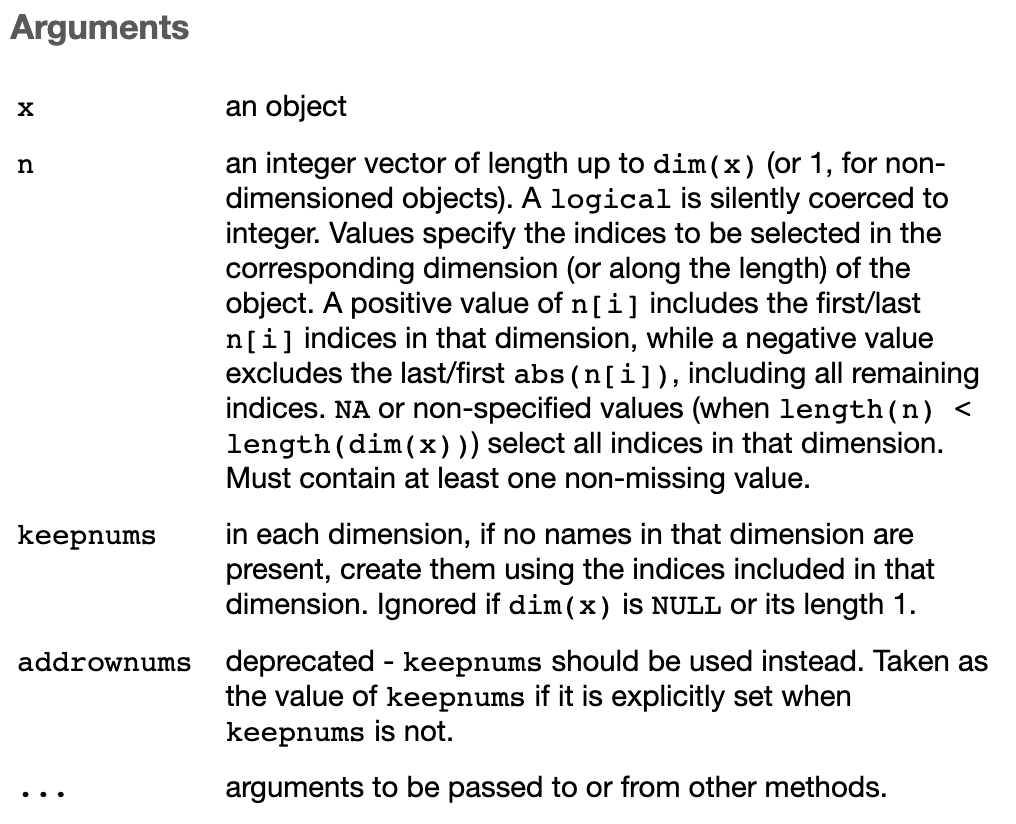
Adding new columns with $ operator
You can add a new column, called log_IgG to df, using the $ operator:
observation_id IgG_concentration age gender slum log_IgG
1 5772 0.3176895 2 Female Non slum -1.146681
2 8095 3.4368231 4 Female Non slum 1.234548
3 9784 0.3000000 4 Male Non slum -1.203973Note, my use of the underscore in the variable name rather than a space. This is good coding practice and make calling variables much less prone to error.
Adding new columns with transform()
We can also add a new column using the transform() function:
Registered S3 method overwritten by 'printr':
method from
knit_print.data.frame rmarkdownTransform an Object, for Example a Data Frame
Description:
'transform' is a generic function, which-at least currently-only
does anything useful with data frames. 'transform.default'
converts its first argument to a data frame if possible and calls
'transform.data.frame'.
Usage:
transform(`_data`, ...)
Arguments:
_data: The object to be transformed
...: Further arguments of the form 'tag=value'
Details:
The '...' arguments to 'transform.data.frame' are tagged vector
expressions, which are evaluated in the data frame '_data'. The
tags are matched against 'names(_data)', and for those that match,
the value replace the corresponding variable in '_data', and the
others are appended to '_data'.
Value:
The modified value of '_data'.
Warning:
This is a convenience function intended for use interactively.
For programming it is better to use the standard subsetting
arithmetic functions, and in particular the non-standard
evaluation of argument 'transform' can have unanticipated
consequences.
Note:
If some of the values are not vectors of the appropriate length,
you deserve whatever you get!
Author(s):
Peter Dalgaard
See Also:
'within' for a more flexible approach, 'subset', 'list',
'data.frame'
Examples:
transform(airquality, Ozone = -Ozone)
transform(airquality, new = -Ozone, Temp = (Temp-32)/1.8)
attach(airquality)
transform(Ozone, logOzone = log(Ozone)) # marginally interesting ...
detach(airquality)Adding new columns with transform()
For example, adding a binary column for seropositivity called seropos:
| observation_id | IgG_concentration | age | gender | slum | log_IgG | seropos |
|---|---|---|---|---|---|---|
| 5772 | 0.3176895 | 2 | Female | Non slum | -1.1466807 | FALSE |
| 8095 | 3.4368231 | 4 | Female | Non slum | 1.2345475 | FALSE |
| 9784 | 0.3000000 | 4 | Male | Non slum | -1.2039728 | FALSE |
| 9338 | 143.2363014 | 4 | Male | Non slum | 4.9644957 | TRUE |
| 6369 | 0.4476534 | 1 | Male | Non slum | -0.8037359 | FALSE |
| 6885 | 0.0252708 | 4 | Male | Non slum | -3.6781074 | FALSE |
Creating conditional variables
One frequently used tool is creating variables with conditions. A general function for creating new variables based on existing variables is the Base R ifelse() function, which “returns a value depending on whether the element of test is TRUE or FALSE or NA.
Conditional Element Selection
Description:
'ifelse' returns a value with the same shape as 'test' which is
filled with elements selected from either 'yes' or 'no' depending
on whether the element of 'test' is 'TRUE' or 'FALSE'.Usage:
ifelse(test, yes, no)
Arguments:
test: an object which can be coerced to logical mode.
yes: return values for true elements of 'test'.
no: return values for false elements of 'test'.Details:
If 'yes' or 'no' are too short, their elements are recycled.
'yes' will be evaluated if and only if any element of 'test' is
true, and analogously for 'no'.
Missing values in 'test' give missing values in the result.Value:
A vector of the same length and attributes (including dimensions
and '"class"') as 'test' and data values from the values of 'yes'
or 'no'. The mode of the answer will be coerced from logical to
accommodate first any values taken from 'yes' and then any values
taken from 'no'.Warning:
The mode of the result may depend on the value of 'test' (see the
examples), and the class attribute (see 'oldClass') of the result
is taken from 'test' and may be inappropriate for the values
selected from 'yes' and 'no'.
Sometimes it is better to use a construction such as
(tmp <- yes; tmp[!test] <- no[!test]; tmp)
, possibly extended to handle missing values in 'test'.
Further note that 'if(test) yes else no' is much more efficient
and often much preferable to 'ifelse(test, yes, no)' whenever
'test' is a simple true/false result, i.e., when 'length(test) ==
1'.
The 'srcref' attribute of functions is handled specially: if
'test' is a simple true result and 'yes' evaluates to a function
with 'srcref' attribute, 'ifelse' returns 'yes' including its
attribute (the same applies to a false 'test' and 'no' argument).
This functionality is only for backwards compatibility, the form
'if(test) yes else no' should be used whenever 'yes' and 'no' are
functions.References:
Becker, R. A., Chambers, J. M. and Wilks, A. R. (1988) _The New S
Language_. Wadsworth & Brooks/Cole.See Also:
'if'.Examples:
x <- c(6:-4)
sqrt(x) #- gives warning
sqrt(ifelse(x >= 0, x, NA)) # no warning
## Note: the following also gives the warning !
ifelse(x >= 0, sqrt(x), NA)
## ifelse() strips attributes
## This is important when working with Dates and factors
x <- seq(as.Date("2000-02-29"), as.Date("2004-10-04"), by = "1 month")
## has many "yyyy-mm-29", but a few "yyyy-03-01" in the non-leap years
y <- ifelse(as.POSIXlt(x)$mday == 29, x, NA)
head(y) # not what you expected ... ==> need restore the class attribute:
class(y) <- class(x)
y
## This is a (not atypical) case where it is better *not* to use ifelse(),
## but rather the more efficient and still clear:
y2 <- x
y2[as.POSIXlt(x)$mday != 29] <- NA
## which gives the same as ifelse()+class() hack:
stopifnot(identical(y2, y))
## example of different return modes (and 'test' alone determining length):
yes <- 1:3
no <- pi^(1:4)
utils::str( ifelse(NA, yes, no) ) # logical, length 1
utils::str( ifelse(TRUE, yes, no) ) # integer, length 1
utils::str( ifelse(FALSE, yes, no) ) # double, length 1ifelse example
Reminder of the first three arguments in the ifelse() function are ifelse(test, yes, no).
| observation_id | IgG_concentration | age | gender | slum | log_IgG | seropos | age_group |
|---|---|---|---|---|---|---|---|
| 5772 | 0.3176895 | 2 | Female | Non slum | -1.1466807 | FALSE | young |
| 8095 | 3.4368231 | 4 | Female | Non slum | 1.2345475 | FALSE | young |
| 9784 | 0.3000000 | 4 | Male | Non slum | -1.2039728 | FALSE | young |
| 9338 | 143.2363014 | 4 | Male | Non slum | 4.9644957 | TRUE | young |
| 6369 | 0.4476534 | 1 | Male | Non slum | -0.8037359 | FALSE | young |
| 6885 | 0.0252708 | 4 | Male | Non slum | -3.6781074 | FALSE | young |
ifelse example
Let’s delve into what is actually happening, with a focus on the NA values in age variable.
[1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE NA TRUE TRUE TRUE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE
[25] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE
[37] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[49] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[61] TRUE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[73] FALSE TRUE TRUE TRUE NA TRUE TRUE TRUE FALSE FALSE FALSE FALSE
[85] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[97] TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[109] FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE NA TRUE TRUE
[121] NA TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[133] FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE
[145] TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[157] FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE
[169] FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE TRUE
[181] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE
[193] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE
[205] TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[217] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[229] TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[241] FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE
[253] FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[265] TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE
[277] FALSE TRUE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[289] TRUE NA FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[301] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE
[313] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE
[325] TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE TRUE FALSE FALSE
[337] FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE
[349] FALSE NA FALSE FALSE TRUE FALSE FALSE FALSE FALSE TRUE TRUE TRUE
[361] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE
[373] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE
[385] TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[397] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[409] TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[421] FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[433] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[445] FALSE FALSE TRUE TRUE TRUE TRUE NA NA TRUE TRUE TRUE TRUE
[457] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[469] FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[481] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE
[493] TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[505] FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE
[517] TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[529] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[541] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[553] FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[565] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[577] FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[589] FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[601] FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[613] TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[625] FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[637] TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE NA FALSE FALSE FALSE
[649] FALSE FALSE FALSENesting two ifelse statements example
ifelse(test1, yes_to_test1, ifelse(test2, no_to_test2_yes_to_test2, no_to_test1_no_to_test2)).
Let’s use the table() function to check if it worked.
| age/ | middle | old | young | NA |
|---|---|---|---|---|
| 1 | 0 | 0 | 44 | 0 |
| 2 | 0 | 0 | 72 | 0 |
| 3 | 0 | 0 | 79 | 0 |
| 4 | 0 | 0 | 80 | 0 |
| 5 | 0 | 0 | 41 | 0 |
| 6 | 38 | 0 | 0 | 0 |
| 7 | 38 | 0 | 0 | 0 |
| 8 | 39 | 0 | 0 | 0 |
| 9 | 20 | 0 | 0 | 0 |
| 10 | 44 | 0 | 0 | 0 |
| 11 | 0 | 41 | 0 | 0 |
| 12 | 0 | 23 | 0 | 0 |
| 13 | 0 | 35 | 0 | 0 |
| 14 | 0 | 37 | 0 | 0 |
| 15 | 0 | 11 | 0 | 0 |
| NA | 0 | 0 | 0 | 9 |
Note, it puts the variable levels in alphabetical order, we will show how to change this later.
Data Classes
Overview - Data Classes
One dimensional types (i.e., vectors of characters, numeric, logical, or factor values)
Two dimensional types (e.g., matrix, data frame, tibble)
Special data classes (e.g., lists, dates).
class() function
The class() function allows you to evaluate the class of an object.
One dimensional data types
- Character: strings or individual characters, quoted
- Numeric: any real number(s)
- Double: contains fractional values (i.e., double precision) - default numeric
- Integer: any integer(s)/whole numbers
- Logical: variables composed of TRUE or FALSE
- Factor: categorical/qualitative variables
Character and numeric
This can also be a bit tricky.
If only one character in the whole vector, the class is assumed to be character
Here because integers are in quotations, it is read as a character class by R.
Note, instead of creating a new vector object (e.g., x <- c("1", "4", "7")) and then feeding the vector object x into the first argument of the class() function (e.g., class(x)), we combined the two steps and directly fed a vector object into the class function.
Numeric Subclasses
There are two major numeric subclasses
Doubleis a special subset ofnumericthat contains fractional values.Doublestands for double-precisionIntegeris a special subset ofnumericthat contains only whole numbers.
typeof() identifies the vector type (double, integer, logical, or character), whereas class() identifies the root class. The difference between the two will be more clear when we look at two dimensional classes below.
Logical
Reminder logical is a type that only has three possible elements: TRUE and FALSE and NA
Note that when creating logical object the TRUE and FALSE are NOT in quotes. Putting R special classes (e.g., NA or FALSE) in quotations turns them into character value.
Other useful functions for evaluating/setting classes
There are two useful functions associated with practically all R classes:
is.CLASS_NAME(x)to logically check whether or notxis of certain class. For example,is.integeroris.characteroris.numericas.CLASS_NAME(x)to coerce between classesxfrom currentxclass into a another class. For example,as.integeroras.characteroras.numeric. This is particularly useful is maybe integer variable was read in as a character variable, or when you need to change a character variable to a factor variable (more on this later).
Examples is.CLASS_NAME(x)
Examples as.CLASS_NAME(x)
In some cases, coercing is seamless
[1] "1" "4" "7"[1] 1 4 7[1] TRUE FALSE FALSEIn some cases the coercing is not possible; if executed, will return NA
Factors
A factor is a special character vector where the elements have pre-defined groups or ‘levels’. You can think of these as qualitative or categorical variables. Use the factor() function to create factors from character values.
[1] "character"[1] "factor"[1] "middle" "old" "young" Note 1, that levels are, by default, set to alphanumerical order! And, the first is always the “reference” group. However, we often prefer a different reference group.
Note 2, we can also make ordered factors using factor(... ordered=TRUE), but we won’t talk more about that.
Reference Groups
Why do we care about reference groups?
Generalized linear regression allows you to compare the outcome of two or more groups. Your reference group is the group that everything else is compared to. Say we want to assess whether being <5 years old is associated with higher IgG antibody concentrations
By default middle is the reference group therefore we will only generate beta coefficients comparing middle to young AND middle to old. But, we want young to be the reference group so we will generate beta coefficients comparing young to middle AND young to old.
Changing factor reference
Changing the reference group of a factor variable.
- If the object is already a factor then use
relevel()function and therefargument to specify the reference. - If the object is a character then use
factor()function andlevelsargument to specify the order of the values, the first being the reference.
Let’s look at the relevel() help file
Reorder Levels of Factor
Description:
The levels of a factor are re-ordered so that the level specified
by 'ref' is first and the others are moved down. This is useful
for 'contr.treatment' contrasts which take the first level as the
reference.Usage:
relevel(x, ref, ...)
Arguments:
x: an unordered factor.
ref: the reference level, typically a string.
...: additional arguments for future methods.Details:
This, as 'reorder()', is a special case of simply calling
'factor(x, levels = levels(x)[....])'.Value:
A factor of the same length as 'x'.See Also:
'factor', 'contr.treatment', 'levels', 'reorder'.Examples:
warpbreaks$tension <- relevel(warpbreaks$tension, ref = "M")
summary(lm(breaks ~ wool + tension, data = warpbreaks))Let’s look at the factor() help file
Factors
Description:
The function 'factor' is used to encode a vector as a factor (the
terms 'category' and 'enumerated type' are also used for factors).
If argument 'ordered' is 'TRUE', the factor levels are assumed to
be ordered. For compatibility with S there is also a function
'ordered'.
'is.factor', 'is.ordered', 'as.factor' and 'as.ordered' are the
membership and coercion functions for these classes.Usage:
factor(x = character(), levels, labels = levels,
exclude = NA, ordered = is.ordered(x), nmax = NA)
ordered(x = character(), ...)
is.factor(x)
is.ordered(x)
as.factor(x)
as.ordered(x)
addNA(x, ifany = FALSE)
.valid.factor(object)
Arguments:
x: a vector of data, usually taking a small number of distinct
values.levels: an optional vector of the unique values (as character strings) that ‘x’ might have taken. The default is the unique set of values taken by ‘as.character(x)’, sorted into increasing order of ‘x’. Note that this set can be specified as smaller than ‘sort(unique(x))’.
labels: either an optional character vector of labels for the levels (in the same order as ‘levels’ after removing those in ‘exclude’), or a character string of length 1. Duplicated values in ‘labels’ can be used to map different values of ‘x’ to the same factor level.
exclude: a vector of values to be excluded when forming the set of levels. This may be factor with the same level set as ‘x’ or should be a ‘character’.
ordered: logical flag to determine if the levels should be regarded as ordered (in the order given).
nmax: an upper bound on the number of levels; see 'Details'.
...: (in 'ordered(.)'): any of the above, apart from 'ordered'
itself.ifany: only add an ‘NA’ level if it is used, i.e. if ‘any(is.na(x))’.
object: an R object.
Details:
The type of the vector 'x' is not restricted; it only must have an
'as.character' method and be sortable (by 'order').
Ordered factors differ from factors only in their class, but
methods and model-fitting functions may treat the two classes
quite differently, see 'options("contrasts")'.
The encoding of the vector happens as follows. First all the
values in 'exclude' are removed from 'levels'. If 'x[i]' equals
'levels[j]', then the 'i'-th element of the result is 'j'. If no
match is found for 'x[i]' in 'levels' (which will happen for
excluded values) then the 'i'-th element of the result is set to
'NA'.
Normally the 'levels' used as an attribute of the result are the
reduced set of levels after removing those in 'exclude', but this
can be altered by supplying 'labels'. This should either be a set
of new labels for the levels, or a character string, in which case
the levels are that character string with a sequence number
appended.
'factor(x, exclude = NULL)' applied to a factor without 'NA's is a
no-operation unless there are unused levels: in that case, a
factor with the reduced level set is returned. If 'exclude' is
used, since R version 3.4.0, excluding non-existing character
levels is equivalent to excluding nothing, and when 'exclude' is a
'character' vector, that _is_ applied to the levels of 'x'.
Alternatively, 'exclude' can be factor with the same level set as
'x' and will exclude the levels present in 'exclude'.
The codes of a factor may contain 'NA'. For a numeric 'x', set
'exclude = NULL' to make 'NA' an extra level (prints as '<NA>');
by default, this is the last level.
If 'NA' is a level, the way to set a code to be missing (as
opposed to the code of the missing level) is to use 'is.na' on the
left-hand-side of an assignment (as in 'is.na(f)[i] <- TRUE';
indexing inside 'is.na' does not work). Under those circumstances
missing values are currently printed as '<NA>', i.e., identical to
entries of level 'NA'.
'is.factor' is generic: you can write methods to handle specific
classes of objects, see InternalMethods.
Where 'levels' is not supplied, 'unique' is called. Since factors
typically have quite a small number of levels, for large vectors
'x' it is helpful to supply 'nmax' as an upper bound on the number
of unique values.
When using 'c' to combine a (possibly ordered) factor with other
objects, if all objects are (possibly ordered) factors, the result
will be a factor with levels the union of the level sets of the
elements, in the order the levels occur in the level sets of the
elements (which means that if all the elements have the same level
set, that is the level set of the result), equivalent to how
'unlist' operates on a list of factor objects.Value:
'factor' returns an object of class '"factor"' which has a set of
integer codes the length of 'x' with a '"levels"' attribute of
mode 'character' and unique ('!anyDuplicated(.)') entries. If
argument 'ordered' is true (or 'ordered()' is used) the result has
class 'c("ordered", "factor")'. Undocumentedly for a long time,
'factor(x)' loses all 'attributes(x)' but '"names"', and resets
'"levels"' and '"class"'.
Applying 'factor' to an ordered or unordered factor returns a
factor (of the same type) with just the levels which occur: see
also '[.factor' for a more transparent way to achieve this.
'is.factor' returns 'TRUE' or 'FALSE' depending on whether its
argument is of type factor or not. Correspondingly, 'is.ordered'
returns 'TRUE' when its argument is an ordered factor and 'FALSE'
otherwise.
'as.factor' coerces its argument to a factor. It is an
abbreviated (sometimes faster) form of 'factor'.
'as.ordered(x)' returns 'x' if this is ordered, and 'ordered(x)'
otherwise.
'addNA' modifies a factor by turning 'NA' into an extra level (so
that 'NA' values are counted in tables, for instance).
'.valid.factor(object)' checks the validity of a factor, currently
only 'levels(object)', and returns 'TRUE' if it is valid,
otherwise a string describing the validity problem. This function
is used for 'validObject(<factor>)'.Warning:
The interpretation of a factor depends on both the codes and the
'"levels"' attribute. Be careful only to compare factors with the
same set of levels (in the same order). In particular,
'as.numeric' applied to a factor is meaningless, and may happen by
implicit coercion. To transform a factor 'f' to approximately its
original numeric values, 'as.numeric(levels(f))[f]' is recommended
and slightly more efficient than 'as.numeric(as.character(f))'.
The levels of a factor are by default sorted, but the sort order
may well depend on the locale at the time of creation, and should
not be assumed to be ASCII.
There are some anomalies associated with factors that have 'NA' as
a level. It is suggested to use them sparingly, e.g., only for
tabulation purposes.Comparison operators and group generic methods:
There are '"factor"' and '"ordered"' methods for the group generic
'Ops' which provide methods for the Comparison operators, and for
the 'min', 'max', and 'range' generics in 'Summary' of
'"ordered"'. (The rest of the groups and the 'Math' group
generate an error as they are not meaningful for factors.)
Only '==' and '!=' can be used for factors: a factor can only be
compared to another factor with an identical set of levels (not
necessarily in the same ordering) or to a character vector.
Ordered factors are compared in the same way, but the general
dispatch mechanism precludes comparing ordered and unordered
factors.
All the comparison operators are available for ordered factors.
Collation is done by the levels of the operands: if both operands
are ordered factors they must have the same level set.Note:
In earlier versions of R, storing character data as a factor was
more space efficient if there is even a small proportion of
repeats. However, identical character strings now share storage,
so the difference is small in most cases. (Integer values are
stored in 4 bytes whereas each reference to a character string
needs a pointer of 4 or 8 bytes.)References:
Chambers, J. M. and Hastie, T. J. (1992) _Statistical Models in
S_. Wadsworth & Brooks/Cole.See Also:
'[.factor' for subsetting of factors.
'gl' for construction of balanced factors and 'C' for factors with
specified contrasts. 'levels' and 'nlevels' for accessing the
levels, and 'unclass' to get integer codes.Examples:
(ff <- factor(substring("statistics", 1:10, 1:10), levels = letters))
as.integer(ff) # the internal codes
(f. <- factor(ff)) # drops the levels that do not occur
ff[, drop = TRUE] # the same, more transparently
factor(letters[1:20], labels = "letter")
class(ordered(4:1)) # "ordered", inheriting from "factor"
z <- factor(LETTERS[3:1], ordered = TRUE)
## and "relational" methods work:
stopifnot(sort(z)[c(1,3)] == range(z), min(z) < max(z))
## suppose you want "NA" as a level, and to allow missing values.
(x <- factor(c(1, 2, NA), exclude = NULL))
is.na(x)[2] <- TRUE
x # [1] 1 <NA> <NA>
is.na(x)
# [1] FALSE TRUE FALSE
## More rational, since R 3.4.0 :
factor(c(1:2, NA), exclude = "" ) # keeps <NA> , as
factor(c(1:2, NA), exclude = NULL) # always did
## exclude = <character>
z # ordered levels 'A < B < C'
factor(z, exclude = "C") # does exclude
factor(z, exclude = "B") # ditto
## Now, labels maybe duplicated:
## factor() with duplicated labels allowing to "merge levels"
x <- c("Man", "Male", "Man", "Lady", "Female")
## Map from 4 different values to only two levels:
(xf <- factor(x, levels = c("Male", "Man" , "Lady", "Female"),
labels = c("Male", "Male", "Female", "Female")))
#> [1] Male Male Male Female Female
#> Levels: Male Female
## Using addNA()
Month <- airquality$Month
table(addNA(Month))
table(addNA(Month, ifany = TRUE))Changing factor reference examples
[1] "young" "middle" "old" OR
df$age_group_factor <- factor(df$age_group, levels=c("young", "middle", "old"))
levels(df$age_group_factor)[1] "young" "middle" "old" Arranging, tabulating, and plotting the data will reflect the new order
Two-dimensional data classes
Two-dimensional classes are those we would often use to store data read from a file
- a matrix (
matrixclass) - a data frame (
data.frameortibbleclasses)
Matrices
Matrices, like data frames are also composed of rows and columns. Matrices, unlike data.frame, the entire matrix is composed of one R class. For example: all entries are numeric, or all entries are character
as.matrix() creates a matrix from a data frame (where all values are the same class). As a reminder, here is the matrix signature function to help remind us how to build a matrix
matrix(data = NA, nrow = 1, ncol = 1, byrow = FALSE, dimnames = NULL)Note, the first matrix filled in numbers 1-6 by columns first and then rows because default byrow argument is FALSE. In the second matrix, we changed the argument byrow to TRUE, and now numbers 1-6 are filled by rows first and then columns.
Data frame
You can transform an existing matrix into data frames using as.data.frame()
You can create a new data frame out of vectors (and potentially lists, but this is an advanced feature and unusual) by using the data.frame() function. Recall that all of the vectors that make up a data frame must be the same length.
Numeric variable data summary
Data summarization on numeric vectors/variables:
mean(): takes the mean of xsd(): takes the standard deviation of xmedian(): takes the median of xquantile(): displays sample quantiles of x. Default is min, IQR, maxrange(): displays the range. Same asc(min(), max())sum(): sum of xmax(): maximum value in xmin(): minimum value in xcolSums(): get the columns sums of a data framerowSums(): get the row sums of a data framecolMeans(): get the columns means of a data framerowMeans(): get the row means of a data frame
Note, all of these functions have an na.rm argument for missing data.
Numeric variable data summary
Let’s look at a help file for range() to make note of the na.rm argument
Range of Values
Description:
'range' returns a vector containing the minimum and maximum of all
the given arguments.Usage:
range(..., na.rm = FALSE)
## Default S3 method:
range(..., na.rm = FALSE, finite = FALSE)
## same for classes 'Date' and 'POSIXct'
.rangeNum(..., na.rm, finite, isNumeric)
Arguments:
...: any 'numeric' or character objects.na.rm: logical, indicating if ‘NA’’s should be omitted.
finite: logical, indicating if all non-finite elements should be omitted.
isNumeric: a ‘function’ returning ‘TRUE’ or ‘FALSE’ when called on ‘c(…, recursive = TRUE)’, ‘is.numeric()’ for the default ‘range()’ method.
Details:
'range' is a generic function: methods can be defined for it
directly or via the 'Summary' group generic. For this to work
properly, the arguments '...' should be unnamed, and dispatch is
on the first argument.
If 'na.rm' is 'FALSE', 'NA' and 'NaN' values in any of the
arguments will cause 'NA' values to be returned, otherwise 'NA'
values are ignored.
If 'finite' is 'TRUE', the minimum and maximum of all finite
values is computed, i.e., 'finite = TRUE' _includes_ 'na.rm =
TRUE'.
A special situation occurs when there is no (after omission of
'NA's) nonempty argument left, see 'min'.S4 methods:
This is part of the S4 'Summary' group generic. Methods for it
must use the signature 'x, ..., na.rm'.References:
Becker, R. A., Chambers, J. M. and Wilks, A. R. (1988) _The New S
Language_. Wadsworth & Brooks/Cole.See Also:
'min', 'max'.
The 'extendrange()' utility in package 'grDevices'.Examples:
(r.x <- range(stats::rnorm(100)))
diff(r.x) # the SAMPLE range
x <- c(NA, 1:3, -1:1/0); x
range(x)
range(x, na.rm = TRUE)
range(x, finite = TRUE)Numeric variable data summary examples
| observation_id | IgG_concentration | age | gender | slum | log_IgG | seropos | age_group | age_group_factor | |
|---|---|---|---|---|---|---|---|---|---|
| Min. :5006 | Min. : 0.0054 | Min. : 1.000 | Length:651 | Length:651 | Min. :-5.2231 | Mode :logical | Length:651 | young :316 | |
| 1st Qu.:6306 | 1st Qu.: 0.3000 | 1st Qu.: 3.000 | Class :character | Class :character | 1st Qu.:-1.2040 | FALSE:360 | Class :character | middle:179 | |
| Median :7495 | Median : 1.6658 | Median : 6.000 | Mode :character | Mode :character | Median : 0.5103 | TRUE :281 | Mode :character | old :147 | |
| Mean :7492 | Mean : 87.3683 | Mean : 6.606 | NA | NA | Mean : 1.6074 | NA’s :10 | NA | NA’s : 9 | |
| 3rd Qu.:8749 | 3rd Qu.:141.4405 | 3rd Qu.:10.000 | NA | NA | 3rd Qu.: 4.9519 | NA | NA | NA | |
| Max. :9982 | Max. :916.4179 | Max. :15.000 | NA | NA | Max. : 6.8205 | NA | NA | NA | |
| NA | NA’s :10 | NA’s :9 | NA | NA | NA’s :10 | NA | NA | NA |
[1] NA NA[1] 1 15[1] 1.665753Character variable data summaries
Data summarization on character or factor vectors/variables using table()
Cross Tabulation and Table Creation
Description:
'table' uses cross-classifying factors to build a contingency
table of the counts at each combination of factor levels.Usage:
table(...,
exclude = if (useNA == "no") c(NA, NaN),
useNA = c("no", "ifany", "always"),
dnn = list.names(...), deparse.level = 1)
as.table(x, ...)
is.table(x)
## S3 method for class 'table'
as.data.frame(x, row.names = NULL, ...,
responseName = "Freq", stringsAsFactors = TRUE,
sep = "", base = list(LETTERS))
Arguments:
...: one or more objects which can be interpreted as factors
(including numbers or character strings), or a 'list' (such
as a data frame) whose components can be so interpreted.
(For 'as.table', arguments passed to specific methods; for
'as.data.frame', unused.)exclude: levels to remove for all factors in ‘…’. If it does not contain ‘NA’ and ‘useNA’ is not specified, it implies ‘useNA = “ifany”’. See ‘Details’ for its interpretation for non-factor arguments.
useNA: whether to include ‘NA’ values in the table. See ‘Details’. Can be abbreviated.
dnn: the names to be given to the dimensions in the result (the
_dimnames names_).deparse.level: controls how the default ‘dnn’ is constructed. See ‘Details’.
x: an arbitrary R object, or an object inheriting from class
'"table"' for the 'as.data.frame' method. Note that
'as.data.frame.table(x, *)' may be called explicitly for
non-table 'x' for "reshaping" 'array's.row.names: a character vector giving the row names for the data frame.
responseName: the name to be used for the column of table entries, usually counts.
stringsAsFactors: logical: should the classifying factors be returned as factors (the default) or character vectors?
sep, base: passed to ‘provideDimnames’.
Details:
If the argument 'dnn' is not supplied, the internal function
'list.names' is called to compute the 'dimname names' as follows:
If '...' is one 'list' with its own 'names()', these 'names' are
used. Otherwise, if the arguments in '...' are named, those names
are used. For the remaining arguments, 'deparse.level = 0' gives
an empty name, 'deparse.level = 1' uses the supplied argument if
it is a symbol, and 'deparse.level = 2' will deparse the argument.
Only when 'exclude' is specified (i.e., not by default) and
non-empty, will 'table' potentially drop levels of factor
arguments.
'useNA' controls if the table includes counts of 'NA' values: the
allowed values correspond to never ('"no"'), only if the count is
positive ('"ifany"') and even for zero counts ('"always"'). Note
the somewhat "pathological" case of two different kinds of 'NA's
which are treated differently, depending on both 'useNA' and
'exclude', see 'd.patho' in the 'Examples:' below.
Both 'exclude' and 'useNA' operate on an "all or none" basis. If
you want to control the dimensions of a multiway table separately,
modify each argument using 'factor' or 'addNA'.
Non-factor arguments 'a' are coerced via 'factor(a,
exclude=exclude)'. Since R 3.4.0, care is taken _not_ to count
the excluded values (where they were included in the 'NA' count,
previously).
The 'summary' method for class '"table"' (used for objects created
by 'table' or 'xtabs') which gives basic information and performs
a chi-squared test for independence of factors (note that the
function 'chisq.test' currently only handles 2-d tables).Value:
'table()' returns a _contingency table_, an object of class
'"table"', an array of integer values. Note that unlike S the
result is always an 'array', a 1D array if one factor is given.
'as.table' and 'is.table' coerce to and test for contingency
table, respectively.
The 'as.data.frame' method for objects inheriting from class
'"table"' can be used to convert the array-based representation of
a contingency table to a data frame containing the classifying
factors and the corresponding entries (the latter as component
named by 'responseName'). This is the inverse of 'xtabs'.References:
Becker, R. A., Chambers, J. M. and Wilks, A. R. (1988) _The New S
Language_. Wadsworth & Brooks/Cole.See Also:
'tabulate' is the underlying function and allows finer control.
Use 'ftable' for printing (and more) of multidimensional tables.
'margin.table', 'prop.table', 'addmargins'.
'addNA' for constructing factors with 'NA' as a level.
'xtabs' for cross tabulation of data frames with a formula
interface.Examples:
require(stats) # for rpois and xtabs
## Simple frequency distribution
table(rpois(100, 5))
## Check the design:
with(warpbreaks, table(wool, tension))
table(state.division, state.region)
# simple two-way contingency table
with(airquality, table(cut(Temp, quantile(Temp)), Month))
a <- letters[1:3]
table(a, sample(a)) # dnn is c("a", "")
table(a, sample(a), dnn = NULL) # dimnames() have no names
table(a, sample(a), deparse.level = 0) # dnn is c("", "")
table(a, sample(a), deparse.level = 2) # dnn is c("a", "sample(a)")
## xtabs() <-> as.data.frame.table() :
UCBAdmissions ## already a contingency table
DF <- as.data.frame(UCBAdmissions)
class(tab <- xtabs(Freq ~ ., DF)) # xtabs & table
## tab *is* "the same" as the original table:
all(tab == UCBAdmissions)
all.equal(dimnames(tab), dimnames(UCBAdmissions))
a <- rep(c(NA, 1/0:3), 10)
table(a) # does not report NA's
table(a, exclude = NULL) # reports NA's
b <- factor(rep(c("A","B","C"), 10))
table(b)
table(b, exclude = "B")
d <- factor(rep(c("A","B","C"), 10), levels = c("A","B","C","D","E"))
table(d, exclude = "B")
print(table(b, d), zero.print = ".")
## NA counting:
is.na(d) <- 3:4
d. <- addNA(d)
d.[1:7]
table(d.) # ", exclude = NULL" is not needed
## i.e., if you want to count the NA's of 'd', use
table(d, useNA = "ifany")
## "pathological" case:
d.patho <- addNA(c(1,NA,1:2,1:3))[-7]; is.na(d.patho) <- 3:4
d.patho
## just 3 consecutive NA's ? --- well, have *two* kinds of NAs here :
as.integer(d.patho) # 1 4 NA NA 1 2
##
## In R >= 3.4.0, table() allows to differentiate:
table(d.patho) # counts the "unusual" NA
table(d.patho, useNA = "ifany") # counts all three
table(d.patho, exclude = NULL) # (ditto)
table(d.patho, exclude = NA) # counts none
## Two-way tables with NA counts. The 3rd variant is absurd, but shows
## something that cannot be done using exclude or useNA.
with(airquality,
table(OzHi = Ozone > 80, Month, useNA = "ifany"))
with(airquality,
table(OzHi = Ozone > 80, Month, useNA = "always"))
with(airquality,
table(OzHi = Ozone > 80, addNA(Month)))Character variable data summary examples
Number of observations in each category
| Female | Male |
|---|---|
| 325 | 326 |
| Female | Male | NA |
|---|---|---|
| 325 | 326 | 0 |
| middle | old | young | NA |
|---|---|---|---|
| 179 | 147 | 316 | 9 |
| Female | Male |
|---|---|
| 0.499232 | 0.500768 |
| middle | old | young |
|---|---|---|
| 0.2788162 | 0.228972 | 0.4922118 |
| middle | old | young |
|---|---|---|
| 0.2788162 | 0.228972 | 0.4922118 |
Summary
- You can create new columns/variable to a data frame by using
$or thetransform()function - One useful function for creating new variables based on existing variables is the
ifelse()function, which returns a value depending on whether the element of test isTRUEorFALSE - The
class()function allows you to evaluate the class of an object. - There are two types of numeric class objects: integer and double
- Logical class objects only have
TRUEorFALSEorNA(without quotes) is.CLASS_NAME(x)can be used to test the class of an object xas.CLASS_NAME(x)can be used to change the class of an object x- Factors are a special character class that has levels
- There are many fairly intuitive data summary functions you can perform on a vector (i.e.,
mean(),sd(),range()) or on rows or columns of a data frame (i.e.,colSums(),colMeans(),rowSums()) - The
table()function builds frequency tables of the counts at each combination of categorical levels
Acknowledgements
These are the materials we looked through, modified, or extracted to complete this module’s lecture.